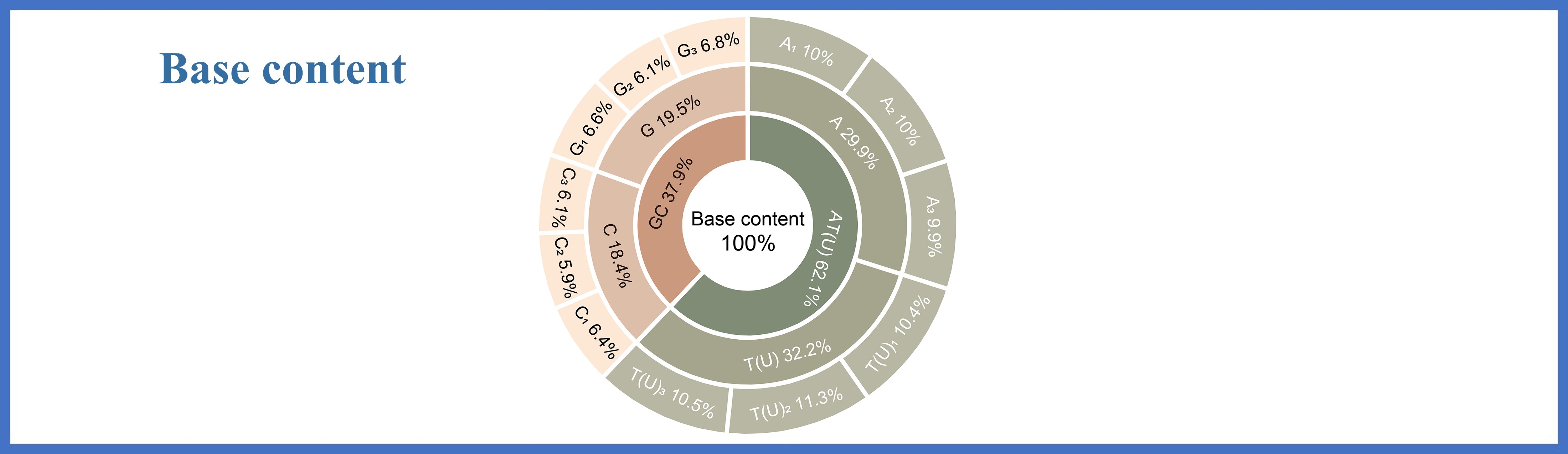
Average nucleotide content at different coding positions of codons
Strains
T(U)
C
A
G
AU
GC
T-1
C-1
A-1
G-1
T-2
C-2
A-2
G-2
T-3
C-3
A-3
G-3
Alpha
32.1
18.3
29.9
19.6
62.0
38.0
33.7
16.8
28.9
20.7
29.4
18.4
30.7
21.5
33.3
19.8
30.2
16.7
Beta
32.1
18.3
29.9
19.6
62.0
38.0
25.9
18.2
30.9
25.0
33.8
21.0
30.3
15.0
36.7
15.9
28.6
18.8
Delta
32.2
18.3
29.9
19.6
62.1
37.9
36.0
15.9
28.5
19.5
26.1
18.4
30.8
24.6
34.4
20.7
30.2
14.7
Gamma
32.2
18.4
29.9
19.6
62.1
37.9
33.7
20.8
30.2
15.3
36.2
16.1
28.6
19.2
26.7
18.1
30.9
24.3
Lambda
32.2
18.3
29.9
19.6
62.1
37.9
36.7
15.8
28.6
18.9
26.0
18.2
30.8
25.0
33.8
21.0
30.3
15.0
Omicron
32.1
18.3
29.9
19.6
62.0
38.0
36.0
16.1
28.6
19.3
26.5
18.2
30.9
24.3
33.8
20.7
30.2
15.3
SARS-CoV-2(SDI)
32.1
18.3
29.9
19.6
62.0
38.0
28.6
18.3
31.0
22.1
33.9
20.0
30.0
16.0
34.0
16.7
28.6
20.7
HCoV-229E
34.5
16.7
27.1
21.7
61.6
38.4
40.2
18.1
25.2
16.4
35.2
14.6
25.4
24.8
28.2
17.4
30.7
23.7
HCoV-HKU1
40.2
13.0
27.8
19.0
62.0
38.0
47.6
9.2
25.5
17.7
30.9
14.6
29.7
24.9
42.1
15.1
28.3
14.5
HCoV-NL63
39.2
14.5
26.3
20.0
65.5
34.5
34.0
14.9
27.2
23.9
40.0
15.6
26.6
17.8
43.5
12.9
25.2
18.4
HCoV-OC43
35.7
15.2
27.7
21.5
63.4
36.6
36.7
17.6
28.2
17.5
42.2
12.3
24.6
20.9
28.2
15.6
30.2
26.0
MERS-CoV
32.6
20.2
26.2
20.9
58.8
41.2
32.0
19.4
25.3
23.3
30.0
20.0
28.6
21.3
35.8
21.3
24.8
18.2
SARS-CoV
30.7
20.0
28.4
20.9
59.1
40.9
30.0
20.2
28.8
21.0
30.0
20.5
28.6
20.9
32.0
19.3
28.0
20.7
HCoVs(SDII)
32.7
18.5
28.6
20.2
61.3
38.7
30.4
18.9
28.7
22.1
33.8
19.0
28.8
18.4
34.0
17.8
28.2
20.0
Strains
ii
si
sv
R
The proportion of ii in the complete genome sequences(%)
Alpha
29763
24
13
1.8
99.88
Beta
29276
20
11
1.9
99.89
Delta
29611
22
9
2.6
99.90
Gamma
29785
28
17
1.6
99.85
Lambda
29783
7
2
3.3
99.97
Omicron
29033
16
5
3.4
99.93
SARS-CoV-2(SDI)
29714
26
14
1.8
99.87
HCoV-229E
25326
1207
618
2.0
93.28
HCoV-HKU1
28955
489
227
2.2
97.59
HCoV-NL63
27250
198
36
5.4
99.15
HCoV-OC43
30399
136
54
2.5
99.38
MERS-CoV
29668
203
158
1.3
98.80
SARS-CoV
28674
623
329
1.9
96.79
HCoVs(SDII)
20815
3049
4058
0.8
74.55
- MEGA
- DNAsp5
Strains
CpG islands
Conserved sites
Variable sites
Parsimony informative sites
Singleton sites
Alpha
7410/29929
27106
2800
1335
1464
Beta
9696/29874
29798
74
51
23
Delta
9694/29822
29530
291
108
183
Gamma
10174/29862
29785
45
0
0
Lambda
10072/29822
29783
13
0
13
Omicron
9638/29838
29663
101
68
33
SARS-CoV-2(SDI)
6782/29999
26752
3171
1555
1607
HCoV-229E
4332/29109
18840
9881
5821
3822
HCoV-HKU1
7160/30435
28213
2000
1743
257
HCoV-NL63
8152/27983
26265
1626
752
837
HCoV-OC43
8518/30896
29189
1624
834
786
MERS-CoV
3080/31014
17005
13897
12037
1843
SARS-CoV
5548/30981
22806
7767
5568
2193
HCoVs(SDII)
412/41729
10517
30162
28584
1400
Strains
Total number of sequences
Total number of sites (excluding sites with gaps / missing data)
Invariable (monomorphic) sites
Variable (polymorphic) sites
Singleton sites
Parsimony informative sites
Two variants
Three variants
Four variants
Two variants
Three variants
Four variants
Ref
1
Alpha
844
26753
24601(91.96%)
2152(8.04%)
1050(48.79%)
15(0.70%)
0
1032(47.96%)
55(2.56%)
0
Beta
8
28418
28357(99.79%)
61(0.21%)
15(24.59%)
0
0
46(75.41%)
0
0
Delta
52
28874
28650(99.22%)
224(0.78%)
123(54.91%)
0
0
98(43.75%)
3(1.34%)
0
Gamma
2
29830
29785(99.85%)
45(0.15%)
45(100.00%)
0
0
0
0
0
Lambda
3
29789
29776(99.96%)
13(0.04%)
13(100.00%)
0
0
0
0
0
Omicron
26
28283
28209(99.74%)
74(0.26%)
27(36.49%)
0
0
47(63.51%)
0
0
SARS-CoV-2(SDI)
936
24757
22639
2118
1006
20
0
1042
50
0
HCoV-229E
35
26522
17757
8765
2955
401
37
3851
1311
210
HCoV-HKU1
41
28724
26782
1942
231
0
0
1664
47
0
HCoV-NL63
74
28482
27108
1374
644
4
0
692
34
0
HCoV-OC43
54
27198
25876
1322
625
6
0
666
24
1
MERS-CoV
421
27696
15489
12207
1352
149
17
6645
3200
844
SARS-CoV
155
28257
21057
7200
1957
96
3
4010
954
180
HCoVs(SDII)
1716
15303
3302
12001
237
9
0
4142
4149
3464
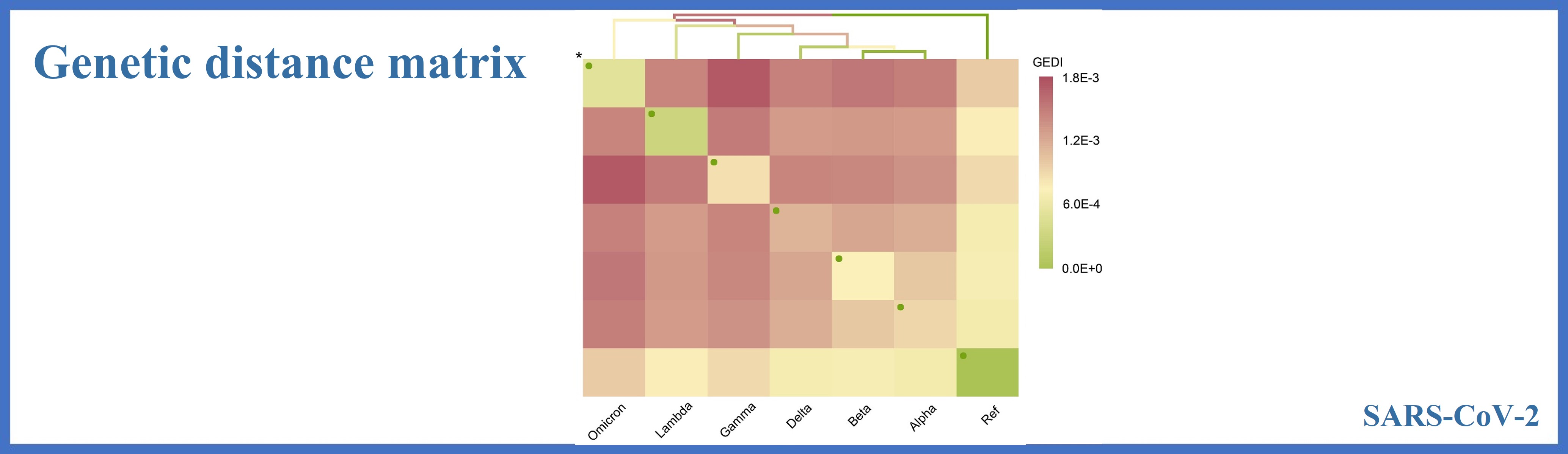
Between group mean distance
Ref
Alpha
Beta
Gamma
Delta
Lambda
Omicron
Ref
0.0000
0.0007
0.0007
0.0009
0.0007
0.0008
0.0010
Alpha
0.0007
0.0009
0.0010
0.0014
0.0012
0.0013
0.0015
Beta
0.0007
0.0010
0.0008
0.0014
0.0012
0.0013
0.0015
Gamma
0.0009
0.0014
0.0014
0.0009
0.0014
0.0015
0.0017
Delta
0.0007
0.0012
0.0012
0.0014
0.0011
0.0013
0.0015
Lambda
0.0008
0.0013
0.0013
0.0015
0.0013
0.0003
0.0014
Omicron
0.0010
0.0015
0.0015
0.0017
0.0015
0.0014
0.0005
SARS-COV-2
HCOV-229E
HCOV-HKU1
HCOV-NL63
HCOV-OC43
MERS-COV
SARS-COV
SARS-COV-2
0.0008
0.7245
0.6518
0.7334
0.6804
0.6034
0.1993
HCOV-229E
0.7245
0.0615
0.7422
0.4821
0.7614
0.7515
0.7421
HCOV-HKU1
0.6518
0.7422
0.0175
0.6188
0.2751
0.6657
0.7015
HCOV-NL63
0.7334
0.4821
0.6188
0.0063
0.6467
0.7635
0.7642
HCOV-OC43
0.6804
0.7614
0.2751
0.6467
0.0043
0.6777
0.7009
MERS-COV
0.6034
0.7515
0.6657
0.7635
0.6777
0.0105
0.6201
SARS-COV
0.1993
0.7421
0.7015
0.7642
0.7009
0.6201
0.0278
Strains
Hd
The weighted values on Phylogenetic trees
π(Nucleotide diversity with JC, PiJC)
The weighted values on Phylogenetic trees
Polymorphism
Ref
\
\
\
\
Hd
π
θ
Alpha
0.99944
\
0.00093
\
0.99960
0.00110
0.01135
Beta
0.96429
\
0.00076
\
0.96400
0.00097
0.00083
Delta
0.99095
\
0.00087
\
0.99800
0.00094
0.00174
Gamma
1.00000
\
0.00113
\
1.00000
0.00151
0.00151
Lambda
1.00000
\
0.00027
\
1.00000
0.00029
0.00029
Omicron
0.97231
\
0.00053
\
0.97200
0.00060
0.00069
SARS-CoV-2(SDI)
0.99949
\
0.00101
\
0.99950
0.00101
0.01191
SARS-CoV-2 (self-contained unit) in SDII
0.99835
1.83500
0.00076
0.00076
\
\
\
HCoV-229E
0.99328
1.32800
0.06089
0.06089
0.99800
0.06642
0.10045
HCoV-HKU1
0.99268
1.26800
0.01742
0.01742
0.99400
0.02458
0.01618
HCoV-NL63
0.99720
1.72000
0.00629
0.00629
0.99800
0.00830
0.01092
HCoV-OC43
0.99223
1.22300
0.00430
0.00430
0.99300
0.00591
0.01017
MERS-CoV
0.99549
1.54900
0.01042
0.01042
0.99800
0.01110
0.09426
SARS-CoV
0.98467
0.46700
0.02752
0.02752
0.99570
0.03118
0.05428
HCoVs(SDII)
0.99909
1.90900
0.24639 (Nucleotide diversity, PiT)
0.24639
0.99910
0.24639
0.18800
.jpg)





 E-mail
E-mail